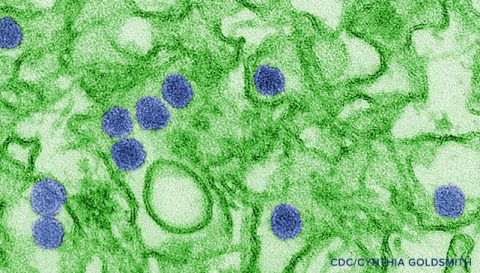
The rapidly developing Zika virus (ZIKV) outbreak has research groups, government agencies, and industry is all striving to develop a response plan to contain and ultimately prevent ZIKV spread. Currently JCVI is working with both private and public sector funders to sequence and analyze historical and current ZIKV strains. Work at JCVI is geared toward developing sensitive ZIKV diagnostics, significantly increasing the number of ZIKV genomic sequences available, and performing cutting-edge analysis on current and future sequence data. We expect these efforts to guide the rational design of ZIKV antivirals and vaccines to treat and prevent ZIKV-induced disease. Here we highlight areas of ongoing ZIKV related work at JCVI. In each area, additional funding would accelerate our efforts to understand and ultimately control ZIKV infection in the human population.
As of October 12, 2016 the Centers for Disease Control and Prevention reported 3,936 cases of ZIKV infection, with two Florida cities identified as the Ground Zero for local transmission.
ZIKV sequencing efforts at JCVI:
- JCVI, through an existing NIH funded grant, is working with the Biodefense and Emerging Infections Research Resources Repository (BEI Resources) to provide high quality sequence data for publically available ZIKV strains. These strains represent a collection of ZIKV isolates, ranging from the initial 1947 isolate from Uganda to 2015 isolates from Puerto Rico, Colombia, and Panama, Mexico, and Honduras. JCVI is providing the gold standard annotated reference sequence for all strains available from BEI and will continue this effort as BEI obtains additional ZIKV isolates.
- Shortly after the recent Zika virus outbreak emerged in eastern Brazil, Dr. Richard Scheuermann and his bioinformatics team at JCVI collaborated with software engineers at Northrop Grumman to develop a custom Zika website portal to provide genomic sequence and other data about Zika virus through the public Virus Pathogen Resource (ViPR). As of September 2016, the ViPR Zika portal contains 389 genomic and 2399 protein sequences representing the three major Zika lineages – East African, West African and Asian. To support comparative genomics analysis to investigate the evolution of virulence in the newly emerging outbreak isolates, Scheuermann’s group developed an algorithm for predicting the proteolytic cleavage sites that generate Zika mature peptides, and applied this method to produce a comprehensive record of all predicted mature peptides for all Zika genomic sequence in the ViPR database.
- JCVI is currently working with collaborators in Colombia and Nicaragua to collect sera from patients suspected to harbor ZIKV and to sequence the viral genome from these patients.
- JCVI was recently awarded NIH supplemental funding to work with Sanofi-Pasteur to screen and sequence human samples suspected to be positive for ZIKV. The majority of samples, provided by Sanofi, are from children and adolescents from the Americas and the South Pacific where mosquito transmitted viruses are common. Over the upcoming year, JCVI anticipates screening both retrospective and prospective human serum samples for ZIKV, with the assumption that many of these samples are from individuals infected with other viral diseases (e.g. Dengue Virus).
Toward the development of a rapid ZIKV diagnostic:
- Brett Pickett recently received funding from the US Agency for International Development (USAID) to develop a method for detecting antibodies against Zika virus in human patients. A bioinformatics analysis performed previously at JCVI uncovered regions of Flavivirus proteins that differentiate between 10 species of viruses—including Zika. Custom peptide arrays will be constructed to identify immunodominant epitopes in human serum, which we will then optimize as an ELISA-based diagnostic for use in developing countries.
- To distinguish between ZIKV and other viral diseases, we are developing a highly sensitive and specific ZIKV diagnostic PCR assay.
- Our assay is sensitive, and we have demonstrated the ability for the assay to identify ZIKV from diverse geographical regions. Future work seeks to move this technology from the laboratory to the field.
Next generation vaccine technology at JCVI can be applied to ZIKV:
- JCVI has previously coupled synthetic biology for the rapid generation of an Influenza vaccine.
- Currently, JCVI is using both synthetic biology and vaccinology to develop a universal vaccine for the common cold in partnership with Synthetic Genomics (A company founded by Dr. Venter) and private funders
- The established vaccine technology at JCVI and our ability to rapidly identify and sequence ZIKV would allow the institute to pursue novel ZIKV vaccine platforms.
Mosquito genomic sequencing:
- JCVI is currently sequencing the genome of a mosquito that is known to harbor ZIKV and is present in the Americas.
- Determining the genomic sequence of this mosquito will help research groups identify develop targeted approaches to impair ZIKV replication in the mosquito host.
Current efforts to combat Zika virus involve CLIA-approved methods to detect viral genetic material. In addition, there are multiple players currently developing a vaccine including GlaxoSmithKline, Sanofi, and Onovio Pharmaceuticals. Ensuring that any vaccine doesn’t cause any neurodevelopmental problems further complicates these efforts. Vector control departments around the United States are currently spraying to eradicate adult and larval mosquitoes. While these endeavors serve to prevent virus infection and spread through mosquitoes, they have negatively affected bee populations and organic crops—potentially increasing public acceptance of sterile GMO mosquitoes.
One of the key questions that arose as a result of the Zika outbreak in the western hemisphere is if the virus has mutated to become more virulent, causing more severe neurological pathology than previously circulating strains. Comparative genomics analysis using sequences and analysis tools in ViPR has identified both nucleotide and amino acid substitutions in the outbreak lineage that warrant further investigation to determine if they are responsible for the apparent increased virulence of the new outbreak strain. With the detection of mosquito-borne transmission in Puerto Rica and the continental US, there is now a critical need for more funding for further research into the genomic determinants of virulence and for accelerated development of targeted diagnostics, therapeutics and vaccines. Donate today!