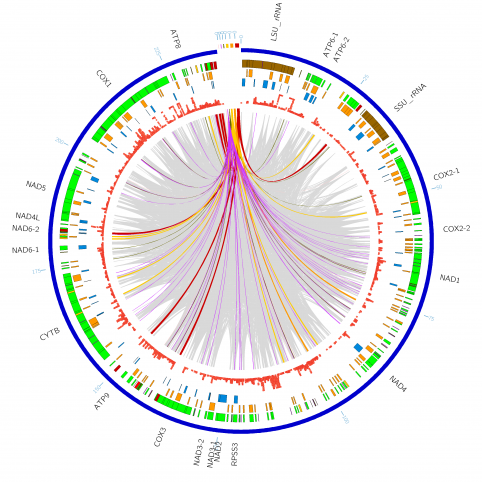
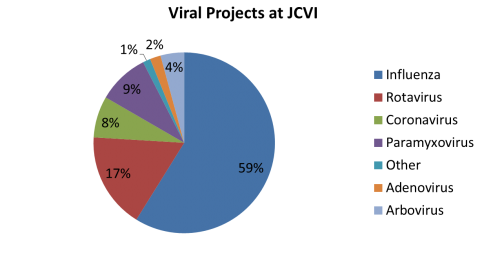
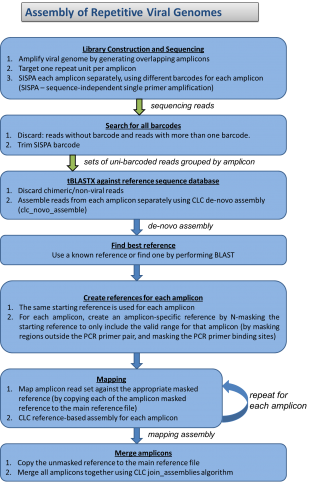
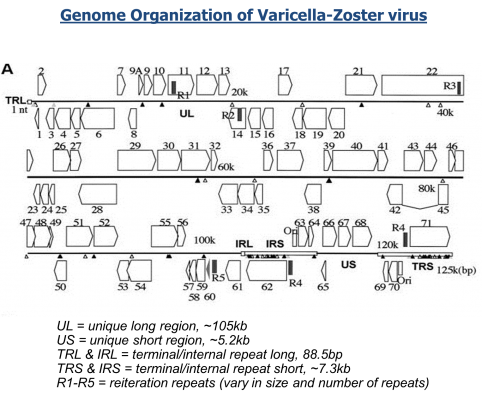
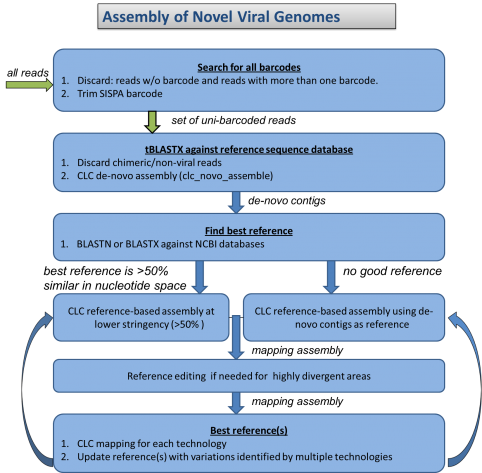
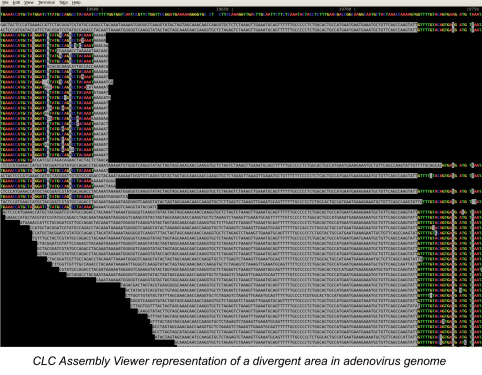
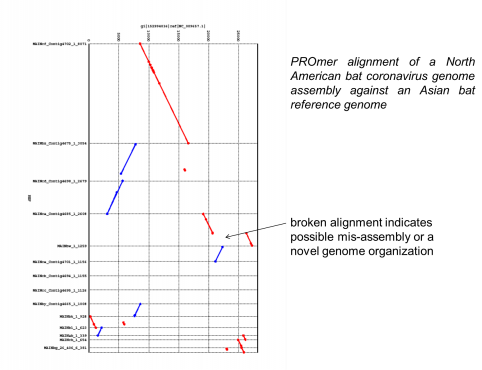
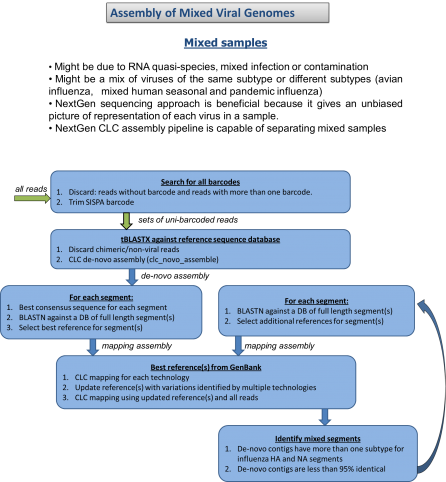
The National Institutes of Health (NIH) and the UK-based Wellcome Trust, in partnership with the African Society of Human Genetics, developed a program to foster genomic and epidemiological research in African scientific institutions. The laboratory and computational infrastructure available to most scientists on the African continent is currently insufficient to keep up with the rapid developments in DNA sequencing technologies and the need to use advanced computationally intensive methods to analyze this data.
Through the H3Africa Consortium, a partnership between NIH and Wellcome Trust, funding has become available to support knowledge development and implementation of genomics-centered research in several African academic institutions. The first scientific paper to come from this effort, Enabeling the Genomic Revolution in Africa, was published in the journal Science in June 2014.
H3Africa Efforts at J. Craig Venter Institute (JCVI)
One of the main initiatives of H3Africa is to foster scientific exchange between US-based partners and their African-based consortium members. JCVI is involved in a number of such partnerships through training and research collaborations.
Tuberculosis Research with Addis Ababa University
Addis Ababa University is the only Ethiopian institution to receive a primary award from NIH under H3Africa. It is based on a collaboration with JCVI. Professor Gobena Ameni of Addis Ababa University and Dr. Rembert Pieper of JCVI developed a proposal on Systems Biology for Molecular Analysis of Tuberculosis in Ethiopia which was initiated earlier this year. The research focuses on genomic variability in M. tuberculosis strains in Ethiopian pastoralist societies and also has an oral microbiome and proteomic biomarker discovery component.
Bioinformatics Training for African Scientists
As part of H3Africa, JCVI is leveraging its recent GCID award, where appropriate, for training of African Scientists. As part of this effort Dr. Andrey Tovchigrechko taught microbiome analysis to graduate students in Ibadan, Nigeria. The workshop was organized by the local H3Africa Bioinformatics Network node. The workshop took place in July, 2014 and comprised of students from Nigeria and other West and Central African countries.

Symposium presenters.

Workshop participants.

The workshop was held at IITA.
During the three day workshop, Dr. Tovchigrechko taught the students launching and controlling computing instances on Amazon cloud, the basics of Python and R programming, MG-RAST Web interface, MG-RAST R package matR and JCVI-developed R code MGSAT. MG-RAST tutorials were provided by one of its developers Andreas Wilke (ANL).
Dr. Tovchigrechko also gave a talk, along with a dozen other speakers, at a one-day symposium at the University of Ibadan that preceded the workshop and included approximately 200 participants. Special thanks go to Nash Oyekanmi, the organizer and manager of the whole event, for his relentless efforts.
Collaborations with University of Cape Town
Also as part of the H3Africa Consortium, Dr. William Nierman from JCVI and Dr. Mark Nicol from the University of Cape Town, South Africa are in collaboration to study the nasopharyngeal microbiome and respiratory disease in African children. Dr. Nierman’s group has conducted a month long in house microbiome training workshop with students from Dr. Nicol’s group.
The focus of the training was to teach students JCVI’s complete microbiome pipeline (including sample preparation, sequencing generation, and final association analysis). The aim of the training collaboration is to ensure that this complete pipeline can be performed at the University of Cape Town, to help build independent and sustainable capacity in this field within South Africa.