Richard H. Scheuermann, Ph.D., who joined JCVI in 2012 from the University of Texas Southwestern as the Director of Bioinformatics, is an accomplished researcher and educator. He and his team apply their deep knowledge in molecular immunology and infectious disease to develop novel computational data mining methods and knowledge representation approaches.

Richard Scheuermann, Ph.D., JCVI’s Director of Bioinformatics
From an early age, Richard was very interested in science and the living world around him. He was a curious child who loved to explore the ponds and fields in his hometown of Warwick, New York. This rural community in upstate New York is covered with dairy farms and apple orchards. He demonstrated an early aptitude for math and science and was fortunate to have talented high school teachers who recognized his potential. Although neither of Richard’s parents were college educated, they encouraged his academic pursuits. When Richard and his father met the high school guidance counselor, Richard told of his intention to attend the Massachusetts Institute of Technology (MIT). The response from the counselor was a resounding, “There is no way you’ll ever get in.” Richard applied to MIT (and only MIT) anyway, and was accepted by early decision.
At MIT Richard intended to pursue a career in chemical engineering (CE) but to his surprise, he found that he loathed the CE classes. While trying to identify a new study path, Richard took a biochemistry class to fulfill a CE requirement. This class would change Richard’s career path. While at MIT, Richard worked in the lab of Annamaria Torrianni-Gorini, Ph.D. and received first hand experience in conducting scientific research. He was inspired by his peers and professors and had found his calling. During this time he also had the privilege to study with Salvador Luria, Ph.D., David Baltimore, Ph.D., David Botstein, Ph.D., and Phil Sharp, Ph.D., all luminaries in their fields. Richard received a B.S. in Life Sciences from MIT in 1981.
Richard went on to complete his Ph.D. in Molecular Biology at the University of California, Berkeley. After completing his doctoral research on bacterial replication fidelity at U.C. Berkeley with Hatch Echols, Ph.D., Richard was offered his own research lab in Europe. He accepted an independent research position at the Basel Institute for Immunology in Switzerland, where he identified the CDP protein as a critical regulator of immunoglobin gene expression and the role of nuclear matrix attachment in transcription regulation.
Although Richard had trained as a molecular biologist, in 1992 he was recruited into the Department of Pathology at the University of Texas Southwestern Medical Center (U.T. Southwestern) in Dallas. Apprehensive at the beginning to find himself in a clinical department, as he had at every previous crossroads, Richard quickly embraced the opportunity before him. He changed his research focus to disease and disease pathogenesis, and he rose to the rank of Professor with tenure. Richard established a robust research program at U.T. Southwestern investigating signal transduction pathways that regulate normal lymphocyte development and function and that induce cell cycle arrest, apoptosis and dormancy in lymphomas. This important work was supported through numerous research grants from the National Institutes of Health, the American Cancer Society, the Texas Higher Education Coordinating Board, and other granting agencies. In the Pathology Department, he also worked on the development and validation of novel diagnostic methods for viruses that mediate chronic infectious disease and for chromosomal translocations that drive leukemia and lymphoma development.
Half way through his U.T. Southwestern career, Richard had found life in the wet lab less and less fulfilling. Through his involvement in several high-throughput research projects Richard realized that it was becoming relatively easy to generate lots of data but more difficult to analyze the information. And so he decided to take a sabbatical year at the San Diego Supercomputer Center to immerse himself into the emerging field of bioinformatics. After his time in San Diego, Richard was drawn into the bioinformatics discipline, and he redirected his career with three research and development proposals funded in rapid succession.
Richard established a very successful bioinformatics program at U.T. Southwestern; however, he was searching for new opportunities to expand on his success. In 2012, the invitation to join JCVI, a “bleeding-edge research institution that valued informatics” could not be ignored.
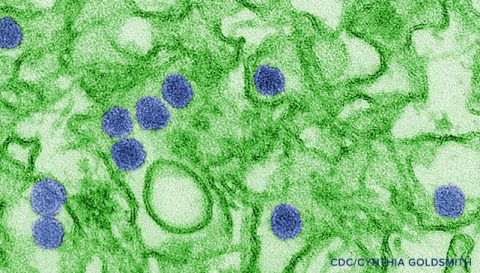
As the Director of Bioinformatics at JCVI, Richard leads a multi-disciplinary team of computational biologists. Richard and his team continue to develop novel computational methods to accelerate data mining and statistical analysis. These methods have been made available to the research community through several public database and analysis resources, including the Influenza Research Database (IRD; www.fludb.org), the Virus Pathogen Resource (ViPR; www.viprbrc.org) and the Immunology Database and Analysis Portal (ImmPort; www.immport.org) supported by the National Institutes of Health. His current research is focused on human pathogenic viruses—how they spread and cause disease. He is a part of the elite community that responds to virus outbreaks, such as the recent Ebola and Enterovirus D68 occurrences. This “real time sorting out” of emerging infectious diseases keeps his still curious New York state of mind engaged and excited about his work.
In his spare time Richard enjoys swimming, soccer and skiing and spending time with his wife, Nancy, and sons, Alex and Derek. Over the past thirteen years, he has studied martial arts, earning his first-degree black belt. As it was in the outskirts of Warwick, Richard continues to explore his new San Diego environment, and is rapidly becoming an avid sailor.
Richard’s commitment and determination to a path, whether on the high seas or in the lab, are unrivaled. Like him, his peers at JCVI are excited to see where his research will take us next.