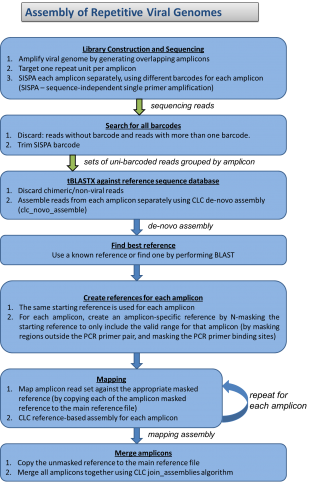
We have all read the stories with concern about the rapid spread of Ebola virus disease (EVD) in Africa. Now, with the first diagnosis of the virus in the United States, it is clear this virus is not under control. If not contained, Ebola poses a significant threat to the African continent and beyond. JCVI is on the front lines of working to better understand this infectious agent. Dr. Reed Shabman, a member of JCVI’s infectious disease team, is seeking to understand why Ebola and Marburg viruses (both are filoviruses) infections result in such severe human disease.
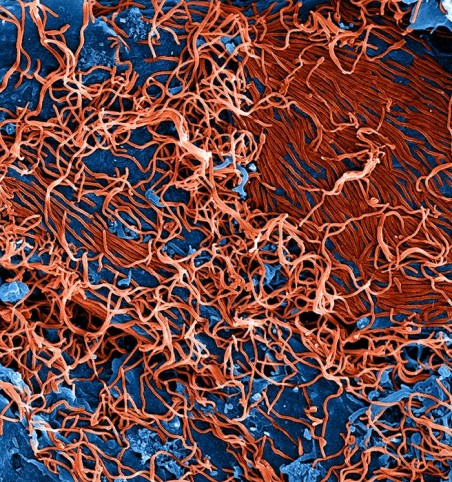
Produced by the National Institute of Allergy and Infectious Diseases (NIAID), under a magnification of 25,000X, this digitally-colorized scanning electron micrograph (SEM) depicts numerous filamentous Ebola virus particles (red) budding from a chronically-infected VERO E6 cell (blue). Image credit: NIAID
During his time as a postdoc at the Icahn School of Medicine at Mount Sinai in New York, Reed helped to develop research platforms designed to understand how Ebola virus mediates its replication, gene expression and evades the immune system. The innovative approaches used at JCVI do not require high level containment facilities and through established collaborations with Biosafety level-4 (BSL-4) labs his group is able to confirm their results in the context of actual Ebola infection.
Some of the ongoing collaborative projects in the group include:
- Determining how Ebola virus evades the host immune system, specifically the innate immune response.
- Employing sequencing platforms to identify previously undescribed aspects of Ebola and Marburg virus RNAs.
- Developing reporter systems to understand how the untranslated regions (UTRs) of Ebola and Marburg virus control their protein production.
This important research seeks to enhance the scientific community’s understanding of Ebola and Marburg virus biology which will aid in our ability to rationally design ways to combat these deadly viruses.
Ebola Background
Ebola has entered the human population before, with the first documented cases occurring in 1976 in areas that are now South Sudan and the Democratic Republic of Congo. Since 1976, there have been approximately 20 outbreaks in central Africa resulting in just over 2300 confirmed cases of Ebola virus disease (EVD).
One unusual aspect of the current Ebola outbreak is that instead of central Africa, this outbreak is occurring in west Africa. Initial cases were reported in February in Guinea. Shortly after these initial reports, EVD spread into Liberia followed by cases in Sierra Leeone and Nigeria. While efforts to control the virus in Nigeria appear to be successful, the number of cases since the first reported case now totals approximately 8000 with almost 4000 fatalities. The numbers from this single outbreak are larger than all other previous outbreaks combined.
Ebola virus was identified almost 40 years ago; however, there are still no approved vaccines or antiviral approaches beyond supportive care. There are promising therapies and vaccines on the horizon, but a fundamental understanding of how the virus interacts with human host is critical to advance the progress of treating the deadly disease.