In the early 2000s, JCVI researchers pioneered in the exploration of the human microbiome, the community of microbes that live in and on the human body. Originally while at The Institute for Genomic Research (TIGR, now part of JCVI) Drs. Craig Venter and Hamilton Smith were awarded a grant from DARPA to examine the microbes found in the human gut. This work was carried out by researchers at JCVI and published in 2006 in Science. While this team had previously published 16S surveys of the human body, this paper in which the researchers found more than 60,000 microbial genes was the first metagenomic description of microbes resided anywhere on the human body. Ten years since this seminal publication, our scientists continue to pave the way for a broader understanding of these vast microbial populations.
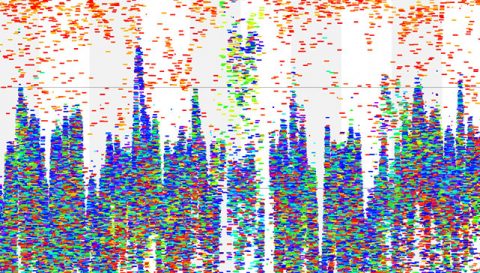
Visualization of ocean microbial data collected on JCVI’s Global Ocean Sampling Expedition (GOS). The Sorcerer II circumnavigated the globe for more than two years, covering a staggering 32,000 nautical miles, visiting 23 different countries and island groups on four continents.
On May 13, 2016, Drs. Craig Venter and Karen Nelson were present at the White House for the launch of the National Microbiome Initiative (NMI). The NMI will invest $121 million in new microbiome studies in fiscal years 2016 and 2017. The goals of the project are to supplement fundamental research, develop new technologies and engage more people in this area of research.
Today we know that the human body is host to more than 1 trillion microbes. Thanks to continued advances in genome sequencing technologies and metagenomic analysis JCVI scientists are providing a deeper understanding of these microbes across a variety of fields. JCVI researchers know that translating the role of the microbiome in the development of health and disease in humans is essential. We believe that eventually the screening of the human microbiome will be a routine part of medical care, leading to prescribed diets and preventive measures personalized to an individual.
JCVI currently has several dozen microbiome studies underway. In this issue of Amplifier, we are highlighting some of our most exciting and cutting edge work unlocking the mysteries of the human microbiome.
The Effects of Long-Term Space Travel on the Microbiome of Astronauts
On March 1, the world celebrated the safe return of NASA astronaut Scott Kelly after 340 days in space. Researchers are fascinated to learn more about the impact of long-term space travel on the human body, and JCVI scientists are excited to be a part of the process. During a mission to space, astronauts are subject to many stressful conditions (g-forces, radiation, microgravity, anxiety, etc.) that can have a negative impact on their health. For example, astronauts lose muscle mass, bone density, and experience a wide range of health problems with everything from their vision to their gastrointestinal tract. Several studies have demonstrated that space travel also affects the astronauts’ immune systems (for example the reactivation of latent viruses like Herpes Simplex Virus 1 and Epstein Bar virus) and have shown some evidence suggesting that stool microbes change after space flight.
JCVI researchers want to determine how the composition of the astronauts’ microbiome changes during long-term space missions (six or more months), and to evaluate potential risks to astronaut health from changes in the microbiome. We are also interested in how the microbiome of astronauts interacts with other factors such as the microbial communities that inhabit the International Space Station (ISS). To accomplish this, we will monitor the astronauts’ health status, environmental stress, and exposure to space conditions. The skin, tongue, nose and gut of each astronaut will be sampled at multiple time points before, during, and after the mission to the ISS. By sampling the microbiome of astronauts on earth while in peak physical health and during subsequent space flight, we will be able to define signatures of human response to a variety of relevant aspects of space travel. Astronauts will also sample different surfaces and the water supply during their stay at the ISS to correlate crew microbiomes with the microbes living at the ISS. We will also assess changes in the astronauts’ immune function and stress levels throughout the mission by analyzing their saliva and blood for metabolic markers. Finally, we will correlate the microbiome and immune function data collected with other measured metadata including astronaut health and hygiene as well as environmental factors such as temperature, humidity and environmental factors.
This research program is being led by Dr. Hernan Lorenzi.
The Gut Microbiome and Human Evolution
Who we are, where we come from and how we came to be as we are, are questions that have always fascinated biologists. The reasons to answer these questions are multiple, but one critical aspect centers on understanding what makes us human. To start addressing these issues JCVI scientists are exploring the gut microbiome of non-human primates, our closest living relatives, and of populations that most faithfully reflect the lifestyles of early hominids: hunter-gatherers. The goal of this project is to establish an evolutionary baseline to shed light on the host-microbe factors that impacted health and disease in modern and western human populations.
Our scientists have shown, in several recent publications, that the gut microbiome of wild gorillas, is strongly shaped by the external environment, namely by diet. Specifically, we showed that gut microbes adapt to different dietary stimuli, probably providing gorillas with energetic plasticity when preferred feeding resources are seasonally and temporally absent. Interestingly, we also suggested that, in conditions in which gorillas exploit high-energy diets, their gut microbiomes resembles those of humans. This fact has critical implications to understanding the evolutionary origins of obesity and inflammation in modern human populations from a microbe perspective. Along these lines, our most recent publication on the gut microbiome of central African hunter-gatherers, traditional agriculturalists and western humans shows evidence that transitions to agriculture and industrialization, and giving up hunting and gathering could have radically changed our gut microbiomes for good. This observation is vital considering that traditional hunter-gatherers, whose microbiomes resemble those of wild gorillas, do not show symptoms of modern inflammatory disease. These observations highlight the potential impact of gut microbes in human evolution.
The research team consists of Drs. Andres Gomez and Karen Nelson.
Solving Crimes with Your Microbial Signature
In January 2016, JCVI received a two-year, $962,500 award from the United States Department of Justice to design and build an open-access microbiome database for the forensic science community. The Forensic Microbiome Database (FMD), the first of its kind will be populated with several thousand microbiome datasets and associated metadata available from the public domain. The database will be based on established procedures for database development designed at the JCVI, incorporating expansive sets of data and metadata that relate to forensic evidence.
The goals of this project are to: provide a host location and continuous monitoring of the database; define well-structured standard operating procedures for data generation and searching against and uploading data into the FMD; and test the utility of the FMD by sequencing a range of samples obtained geographically for querying and proof of concept against the database. The foundation of this project will serve for future enhancements of the FMD and utility for forensic casework. The research team expects this will become the community resource for analysis of microbiome data and for attributing weight to microbial forensic evidence.
The research team consists of Rhonda Roby, Lauren Brinkac, Toby Clarke, Andres Gomez, Karen Nelson, Harinder Singh, and Shibu Yooseph,
Using the Microbiome to Advance Wound Therapies
Chronic wounds are wounds that fail to heal after 4 months of proper wound care and management. It is a major public healthcare burden that affects an estimated 1% of the US population and costs $25 billion per year. Common chronic wounds are leg, foot, and pressure ulcers occur in adults especially the elderly with diabetes, vascular diseases, or specific body locations under prolonged pressure. According to the Centers for Disease Control and Prevention, approximately 12% of U.S. adults with diabetes had a history of foot ulcer and 11% of U.S. nursing home residents had pressure ulcers. In addition to the economic burden, from the perspective of patients chronic wounds can also lead to loss of function (e.g. amputation), decreased quality of life, and increased rate of mortality.
At JCVI we are interested in deciphering not only the microbial communities present in chronic wounds but also their potential impacts and relationship with the wound healing outcome, for working towards more effective clinical strategies of wound healing. In collaboration with George Washington University, we are conducting a study to analyze chronic wounds. Samples are selected from patients enrolled in the Wound Etiology and Healing biospecimen and data repository (WE-HEAL). We will analyze the chronic wound microbiome at the molecular level, and attempt to identify biological indicators that can be used to predict the healing outcome to further advance wound therapies and management.
This project is being led by Dr. Agnes Chan.
Metagenomic Epidemiology of Antibiotic Resistance in Infectious Diarrhea
Genes that encode antimicrobial resistance (AMR) to antibiotics have been detected in environmental, insect, human and animal metagenomes and the sum of these are known as “resistomes.” While metagenomic datasets have been mined to characterize the healthy human gut resistome, directed metagenomic sequencing has not been used to examine the spread of AMR. Especially in developing countries where sanitation is poor, diarrhea and enteric pathogens likely serve to disseminate AMR elements of clinical significance. Unregulated use of antibiotics further exacerbates the problem by selection for acquisition of resistance. This is exemplified by recent reports of multiple AMR in Shigella strains in India, in Escherichia coli in India and Pakistan, and in nontyphoidal Salmonella (NTS) in South-East Asia.
Sarah Highlander, Ph.D. and her team are characterizing the microbial composition and its component AMR transfer elements (such as plasmids and transposons) by metagenomic sequencing of stool samples from pediatric patients from Colombia who are suffering from diarrhea. Our goal is to assess whether groups of species/strains associate with specific mobile genetic elements and whether their presence is enhanced or amplified in diarrheal microbiomes. This work could potentially identify clonal complexes with enhanced resistance and potential pathogenesis.
For more information on how you can support or human microbiome research program at JCVI, please contact [email protected].







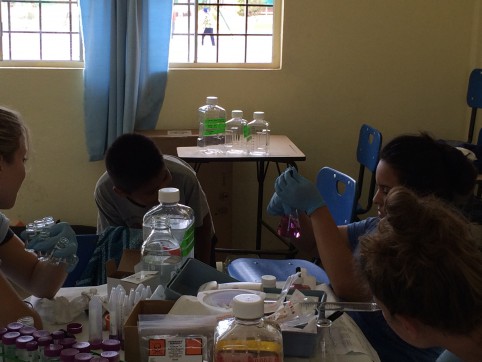




 Pictures from Antarctic Expedition
Pictures from Antarctic Expedition