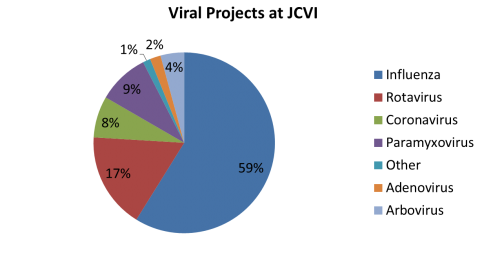
JCVI viral projects are supported by the NIAID Genomic Sequencing Center for Infectious Disease (GSCID). The viral sequencing and finishing pipeline at JCVI combines next generation sequencing technologies with automated data processing. This allowed us to complete over 1,800 viral genomes in the last 12 months, and almost 8,800 genomes since 2005.
Our NextGen pipeline, which utilizes SISPA-generated libraries with Roche/454 and Illumina sequencing, enables us to complete a wide variety of viral genomes including challenging samples. Automated assembly pipeline employs CLCbio command-line tools and JCVI cas2consed, a cas to ace assembly format conversion tool. Our complimentary Sanger pipeline software is currently being integrated with the NextGen pipeline. This will improve our data processing and will allow us to use validation software (autoTasker) more efficiently.
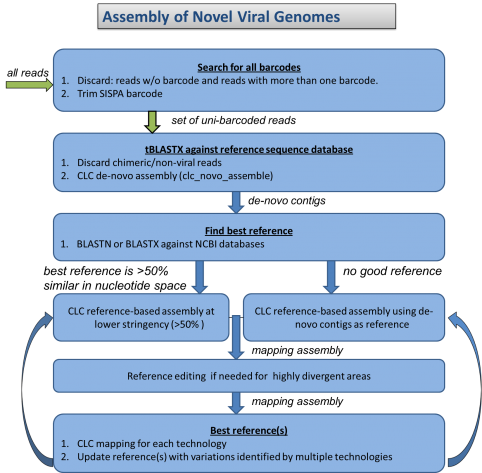
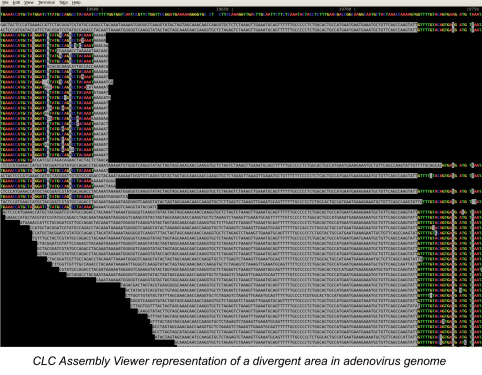
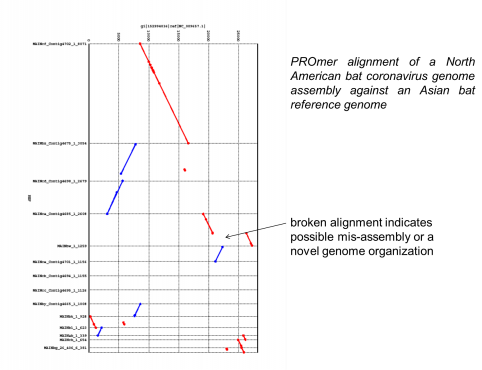
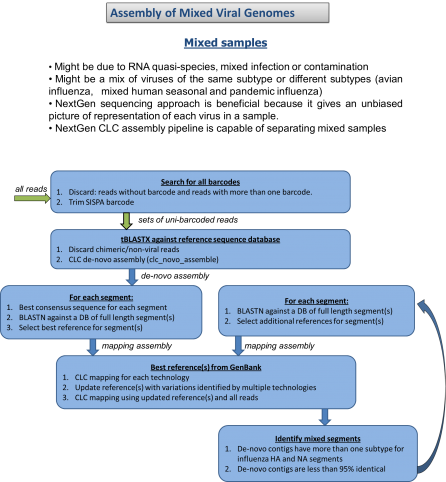
During the past year we have found that novel viruses, repetitive genomes, and mixed infection samples could not be easily integrated with our high-throughput assembly pipeline. We have developed an assembly and finishing process that utilizes components of the high-throughput pipeline and combines them with manual reference selection and editing. Using this approach we completed novel adenovirus genomes and mixed-infection avian influenza genomes, and improved assemblies of previously unknown arbovirus genomes. We are currently working on optimizing and automating this new pipeline.
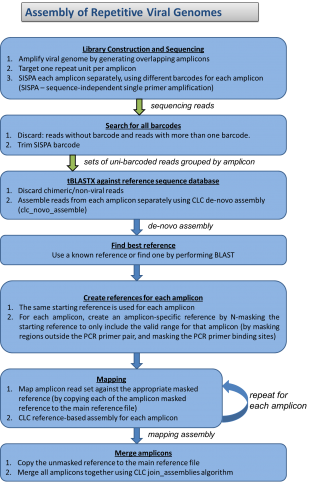
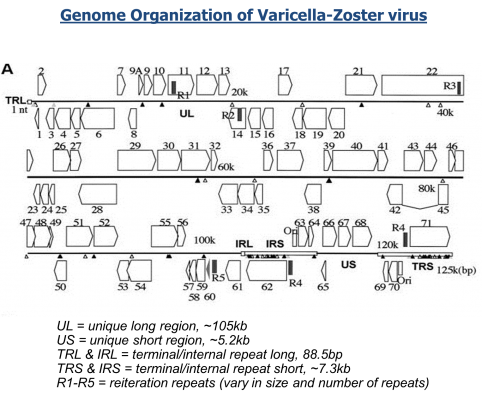
Repetitive genomes have long been known to present great challenges during assembly and finishing. We are presenting a new approach to assembly and finishing of repetitive varicella genome that is based on separating it into overlapping PCR amplicons followed by merging sequenced amplicons during assembly.
To streamline our viral pipelines, we have fully integrated them with JCVI’s LIMS and JIRA Workflow Management to create a semi-automated tracking interface that follows the progress of viral samples from acquisition through to NCBI submission. This allows us to process a large volume of samples with limited manual interaction and, at the same time, gives us flexibility to work on challenging and novel genomes.
Acknowledgements
The JCVI Viral Genomics Group is supported by federal funds from the National Institute of Allergy and Infectious Disease, the National Institutes of Health, and the Department of Health and Human Services under contracts no. HHSN272200900007C.
Bat coronavirus project is collaboration with Kathryn Holmes and Sam Dominguez, University of Colorado Medical Center.
The authors would like to thank members of the Viral Genomics and Informatics group at JCVI.
References
Viral genome sequencing by random priming methods. Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. BMC Genomics. 2008 Jan 7;9:5A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J.
Note
This post is based on a poster by Nadia Fedorova, Danny Katzel, Tim Stockwell, Peter Edworthy, Rebecca Halpin, and David E. Wentworth.