Corbata: CORe microBiome Analysis Tools
Overview
Corbata is a set of R scripts that were used to generate the plots and some statistics for the manuscript, "Analyses of the Stability and Core Taxonomic Memberships of the Human Microbiome", by Li K, Bihan M, and Methé BA.
This document serves as a general introduction to the suite of tools. The software is available on SourceForge.
Corbata is used to analyze the core members across the microbiome of a group of samples (cohort). When you have taxonomic profiles for each sample, for example a table of counts from RDP or OTUs after 16S rRNA sequencing, it is possible to elect who key members are and get a visualization of how these members are spread across your cohort by examining 2 parameters: abundance and ubiquity (prevalence). The tools included in the Corbata suite include: Ubiquity-Abundance plot (for major and minor core), Ubiquity-Ubiquity plot (comparing 2 cohorts), Taxonomic Variance plots, and bootstrapping Core Counts. The power behind using Corbata is its sensitivity to low abundance taxa. Instead of discarding low abundance taxa in core analyses, Corbata tries to expose them.
Current functionality includes:
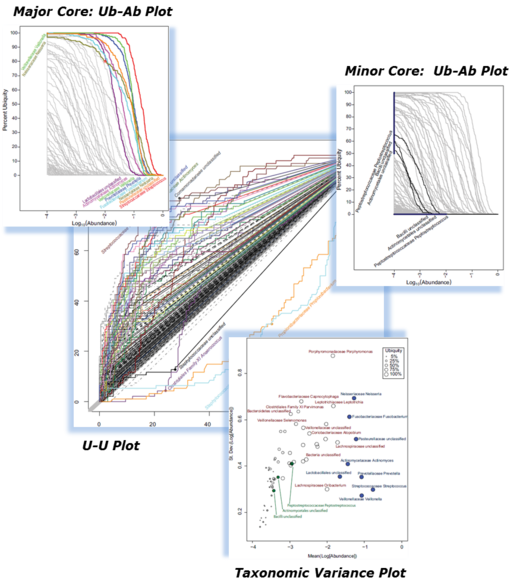
- Ubiquity-Abundance (Ub-Ab) Plot: This plot illustrates the continuous relationship between the abundance and ubiquity of each taxa in your cohort.
- Primary Core: By setting a ubiquity and abundance cutoff, it is possible to identify what the core taxonomic members in your cohort are.
- Minor core: By seeking out low abundance taxa with moderately high ubiquity, it is possible to identify what we call the "minor" core. These taxon may be interesting because they seem to thrive in the background.
- Ubiquity-Ubiquity (U-U) Plot: This plot compares the microbiome between two cohorts.
- Taxonomic Variation: This help to sort out the relationship between mean and variation of abundance for each taxa. Core taxa tend to have low variation.
- Core Numbers: Estimating the confidence intervals around core numbers is critical because of the continuous relationship between abundance and ubiquity. Since there is often not a single pair of parameters that can differentiate core from non-core, it is important to see how "blurry" this boundary is for your dataset.
As our research in metagenomics is quite active, we are constantly improving existing and developing new tools to help identify answers to new questions. As such, the tools provided here are likely to only be a snapshot of the current state of our art. If you have questions or issues with the software, please feel free to contact one of the authors (kli(AT)jcvi.org). We may have an updated version of a script that has not been formally released that we could provide to you. If not, we may try to point you in the best direction to help you make custom modifications yourself.
Sample Output