
Flock: Flow Cytometry Clustering without K
Overview
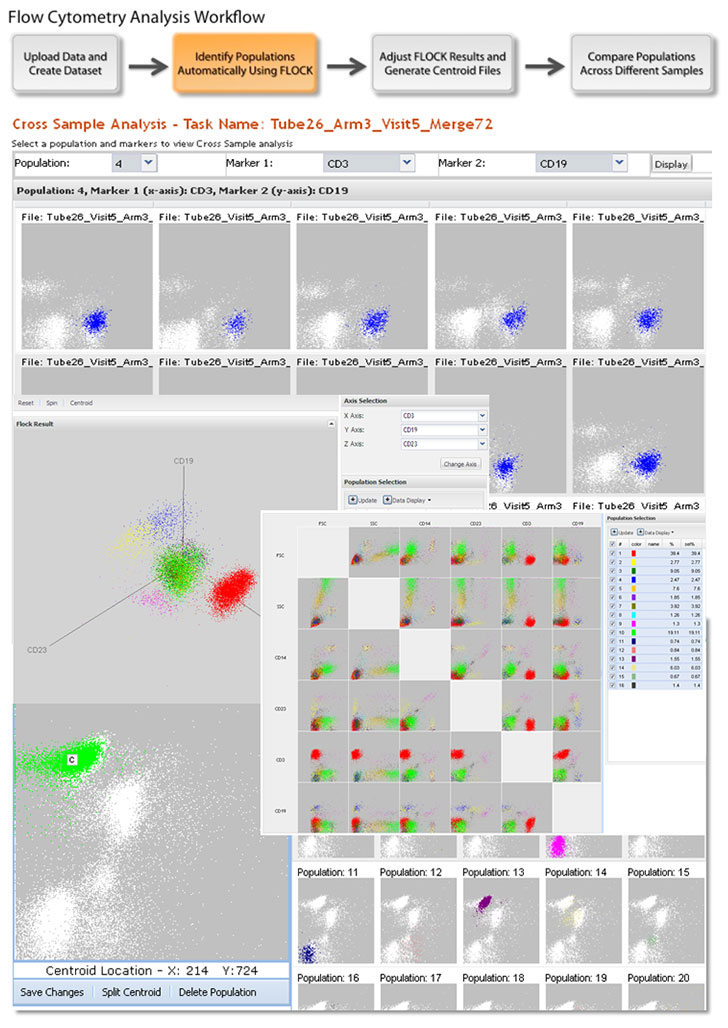
What is FLOCK?
FLOCK (FLOw Clustering without K) is a computational algorithm to identify cell populations within multi-dimensional flow cytometry (FCM) data. It has been implemented as a software system available through the NIAID/DAIT-funded Immunology Database and Analysis Portal.
Why you need FLOCK?
Traditional manual segregation (a.k.a., gating) of cell populations is subjective, labor-intensive, irreproducible, and not scalable to the increasing number of FCM parameters. Computational approaches can reduce human bias and will ultimately replace the manual gating. The use of objective algorithmic approaches like FLOCK should ultimately play an important role in the improvement of clinical diagnosis based on FCM.
What can you do with FLOCK?
- Automated cell population identification
- Result visualization in 2D and 3D
- Statistical analysis of population characteristics
- Centroid-based mapping of cell populations across multiple samples
Links
http://immportflock.sourceforge.net
Publications
Qian Y, Liu Y, Campbell J, Thomson E, Kong YM, Scheuermann RH. FCSTrans: an open source software system for FCS file conversion and data transformation. Cytometry A. 2012 May;81(5):353-6. PMCID: PMC3932304.
Qian Y, Wei C, Lee FE-H, Campbell J, Halliley J, Lee JA, Cai J, Kong YM, Sadat E, Thomson E, Dunn P, Seegmiller AC, Karandikar NJ, Tipton CM, Mosmann T, Sanz I, and Scheuermann RH. Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B, 2010, 78 Suppl 1, S69–82. PMCID: PMC3084630.
Funding
National Institute of Allergy and Infectious Disease (NIAID)
