
Meta-CATS: Metadata-driven Comparative Analysis Tool for Sequences
Overview
What is it?
The metadata-driven comparative analysis tool for sequences (meta-CATS) rapidly identifies sequence variations that significantly correlate between groups of viruses with different attributes.
How do I use it?
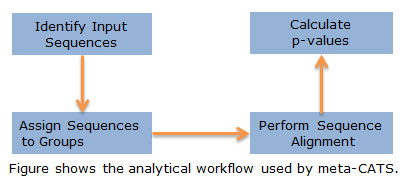
- Go to the meta-CATS page in IRD or ViPR
- Specify the sequences that you want to include in the analysis
- Divide your sequences into groups based on: host species, year or place of isolation, virulence, tropism, etc.
- Verify the groupings
- Click “Run”
What can you do?
- Meta-CATS performs tandem chi-square tests to identify significant positions that vary according to the specified sequence groupings
- The results are helpful in generating hypotheses that can be tested experimentally.
Links
Publications
Pickett BE, Liu M, et al.
Metadata-driven Comparative Analysis Tool for Sequences (meta-CATS): an Automated Process for Identifying Significant Sequence Variations That Correlate With Virus Attributes.
Virology. 2013 Dec 01; 447: 45-51.[more]
Pickett BE, Greer DS, et al.
Virus Pathogen Database and Analysis Resource (ViPR): a Comprehensive Bioinformatics Database and Analysis Resource for the Coronavirus Research Community.
Viruses. 2012 Nov 01; 4: 3209-26.[more]
Pickett BE, Sadat EL, et al.
ViPR: an Open Bioinformatics Database and Analysis Resource for Virology Research.
Nucleic Acids Research. 2012 Jan 01; 40: D593-8.[more]
Funding
National Institute of Allergy and Infectious Disease (NIAID)
