
Variant Classifier
Output from the VariantClassifier
Because the output from the VariantClassifier can be very dense, we provide two file formats: denormalized and normalized. Both are documented in the manual, Manuals/VI_SNP_Classification_File_Formats.pdf and have the same amount of information.
Denormalized Format
The denormalized file format is easier to parse if you plan on reading the information into a downstream script or loading into a database. All the information for each variant exists on a single line, and information can be extracted by column, by splitting lines into columns where tabs are detected.
Normalized Format
The normalized file format is easier to peruse, even with a text editor. However we still suggest using Microsoft Excel, or OpenOffice Calc to help you control the column widths. With either application, read in the file with the text importer with tabs as the field separators. See Examples/human/classify_variants/output.normal. Below is the recommended visualization method with the help of OpenOffice.
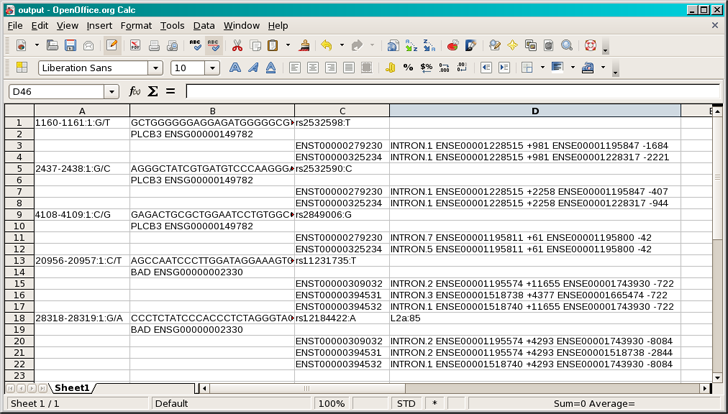
Quick Overview
From the above example, you can see on the first line the variants that were classified, followed by flanking sequence, and the dbSNP ID and any repeats that the variant overlapped. On the next line was the gene that the variant was found in, followed by all the transcripts that the variant was found in. Since there are multiple transcripts associated with the gene, an analysis is performed on each one, hierarchically. Please refer to the manual for a full list of output features.
