Influenza Genome Project
The influenza genome sequencing project (IGSP) was initiated in 2005 to investigate influenza evolution by providing a public data set of complete influenza genome sequences from collections of isolates representing diverse temporal, spatial and species distributions. One of NIAID's primary goals for the project is to dramatically improve the availability of influenza genomic sequence in the public domain. We will sequence the complete genomes of a large collection of human influenza isolates, as well as a select number of avian and other non-human influenza strains important in the evolution of viruses with pandemic potential. The strains will be chosen to represent many subtypes with a wide geographical and chronological distribution. All the data generated will be released to the public domain in accordance with NIAID's data and reagent sharing and release guidelines.
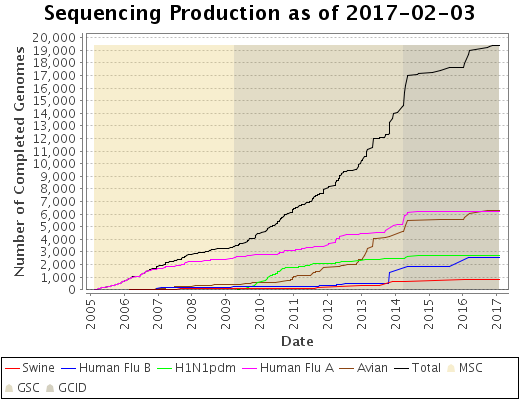
Status Graph
The collection sequencing status is available in real-time here.
There are several questions that need to be answered to better understand the evolution of the influenza virus and help federal and local governments plan responses to inevitable pandemics. These questions include but are not limited to:
- Can next generation deep sequencing technologies be used to better understand the molecular mechanisms central to influenza evolution in response to selection pressures such as: antiviral treatment,emergence in new host species,and pre-existing immunity?
- Are there other influenza strains co-circulating within humans?
- Can the association of genomic sequences with clinical and geographic meta-data reveal genetic determinants responsible for virulence and/or human-to-human transmission?
- What minor populations of virus are present but poorly detected during a given influenza season?
- Can sequencing of influenza viruses present during one period reveal the ancestral strains of a new outbreak and help explain mechanisms of zoonoses?
- Can sequence data be used to predict widespread emergence of antigenic drift variants and aid in vaccine selection?
- Do changes in the untranslated regions of the genome influence pathogenesis,transmission,or zoonosis?
- How does influenza infection influence the microbiome of the host?
- What are the linkages between genomic RNA segments critical to viral fitness?
- Can deep sequencing of a subset of patient samples based on clinical characteristics identify intra-host variation that could serve as an early warning system for the presence of new viral evolutionary trends?
To better answer these and other questions there is a need to generate complete genome information from viruses currently circulating within and between various reservoirs as well as from historical collections. Given the success of the NIAID/JCVI influenza genome sequencing project (IGSP) we propose to lead genomics efforts with a particular focus on complete genome sequencing and the use of bioinformatics tools developed at JCVI as a part of the IGSP to answer the questions posed above. Furthermore, we recognize the importance of using cutting edge deep-sequencing techniques and select sets of samples, particularly those collected directly from the host based on clinical characteristics to understand evolutionary trends with public health significance.
For information about getting your collection sequenced, please visit NIAID's Sequencing Request Process page.
Influenza primer sets are available for download here.
Investigators and Collaborators
Wentworth, David | Director Viral Programs. J.Craig Venter Institute |
Dugan, Vivien | Assistant Professor, J. Craig Venter Institute |
Ghedin, Elodie | JCVI, University of Pittsburgh School of Medicine |
