INPUT FORMAT FOR SIFT INDEL
Reference human genome assembly GRCh37
Format Example: SPACE BASED COORDINATE SYSTEM (comma separated)
Deletion:
7,117180216,117180219,1,/,CFTR;CTT/-;FF/F
7,117199640,117199643,1,/,CFTR;ATC/-;I/-
7,117199640,117199644,1,/,CFTR;frameshift
7,117199646,117199649,1,/,CFTR;TTT/-;F/-;F508
10,50184922,50184925,1,/,WDFY4;TGG/-;W/-
10,102762472,102762475,1,/,GAT/-;D/-
12,121438956,121438959,1,/,rs112974205;HNF1A;ACC/-;T/-
Insertion:
1,43217995,43217995,-1,CCACCA,LEPRE1;insertion;/PP
10,102762472,102762472,1,GCGGCT,insertion;/AA
10,50184922,50184922,1,ACACACACACAC,insertion;/THTH
Reference human genome assembly NCBI36
Format Example: SPACE BASED COORDINATE SYSTEM (comma separated)
10,102760304,102760304,1,GCGGCT,#User comment 1
10,50205013,50205013,1,ACACACACACAC
5,179134934,179134935,1,/,#User comment 2
1,153108866,153108866,1,CTGCTGCTGCTG
11,6368547,6368547,1,GCTGGCGCTGGC
11,65081932,65081932,1,AGCAGC
12,110521161,110521164,1,/
12,116990733,116990736,1,/
12,123453048,123453048,1,CTG
12,131113090,131113090,1,GCA
12,1932613,1932613,1,CTG
Format Description
SIFT Indel input format: [comma separated: chromosome,coordinate,oientation,alleles,user comment(optional)]
Please do not use spaces except in the user comments field
Coordinate System:
SIFT accepts only space-based coordinates for insertion / deletion variants.
The space-based coordinate system counts the spaces before and after bases rather than the bases themselves.
Zero always refers to the space before the first base.
The sequence 'ACGT' has coordinates (0,4) and its subsequence 'CG' has coordinates (1,3) as shown in Example 1 below.
The difference between the start and end coordinates gives the sequence length. Misinterpretation of these
coordinates can easily lead to 'off-by-one' errors. Space-based coordinates become necessary when describing
insertions/deletions and genomic rearrangements.
Example 1:
0 A 1 C 2 G 3 T 4
Orientation:
Use 1 for positive strand and -1 for negative strand. If orientation is not known, use 1 as default.
Alleles:
For Insertion, the begin and end coordinates should be same and the allele should be a string of inserted nucleotides in one of the following formats.
1. ----/ATGC
2. -/ATGC
3. ATGC
For Deletion, the difference between begin and end coordinates should be equal to the length of the deleted string. the allele can either be left blank or be specified in one of the followig formats
1. ATGC/----
2. ATGC/-
3. /
Output Description
Amino Acid Position Change
This column contains the change coordinates within the original protein sequence and the modified protein sequence. For example, the insertion of GCGGCT at location 102760304 of chromosome 10 of Homo Sapiens (represented by input row: 0,102760304,102760304,1,GCGGCT) inserts two additional amino acids Arginine 'R' and Serine 'S' at position 145 to 147 (space based coordinates) in the modified protein sequence. The mismatch between the original and modified sequences is displayed in red.
>ENST00000238965; MISMATCH = 145-145
GPQEQGSPASCFETSPAGHATQASPYHPRACRGGFYLLPVNGFPEEEDNGELRERLGALK
VSPSASAPRHPHKGIPPLQDVPVDAFTPLRIACTPPPQLPPVAPRPLRPNWLLTEPLSRE
HPPQSQIRGRAQSRSRSRSRSRSRSSRGQGKSPGRRSPSPVPTPAPSMTNGRYHKPRKAR
PPLPRPLDGEAAKVGAKQGPSESGTEGTAKEAAMKNPSGELKTVTLSKMKQSLGISISGG
IESKVQPMVKIEKIFPGGAAFLSGALQAGFELVAVDGENLEQVTHQRAVDTIRRAYRNKA
REPMELVVRVPGPSPRPSPSDSSALTDGGLPADHLPAHQPLDAAPVPAHWLPEPPTNPQT
PPTDARLLQPTPSPAPSPALQTPDSKPAPSPRIP
>ENST00000238965; MISMATCH = 145-147
GPQEQGSPASCFETSPAGHATQASPYHPRACRGGFYLLPVNGFPEEEDNGELRERLGALK
VSPSASAPRHPHKGIPPLQDVPVDAFTPLRIACTPPPQLPPVAPRPLRPNWLLTEPLSRE
HPPQSQIRGRAQSRSRSRSRSRSRSrsSRGQGKSPGRRSPSPVPTPAPSMTNGRYHKPRK
ARPPLPRPLDGEAAKVGAKQGPSESGTEGTAKEAAMKNPSGELKTVTLSKMKQSLGISIS
GGIESKVQPMVKIEKIFPGGAAFLSGALQAGFELVAVDGENLEQVTHQRAVDTIRRAYRN
KAREPMELVVRVPGPSPRPSPSDSSALTDGGLPADHLPAHQPLDAAPVPAHWLPEPPTNP
QTPPTDARLLQPTPSPAPSPALQTPDSKPAPSPRIP
Indel location
This percentage indicates the approximate location of the indel in the protein.
For example, a value less than 50% means that the indel is located in the first half of the protein and is close to the amino terminus, whereas a number greater than 50% means that the indel is closer to the carboxy terminus.
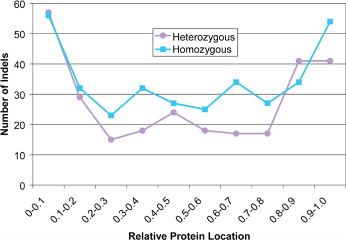
On the x-axis is the relative protein location of the coding indel, which is the first amino acid position of the indel divided by the protein length. A relative protein location near zero indicates that the indel is located near the N-terminus of the protein and a relative protein location near one indicates that the indel is located near the C-terminus of the protein.
Plotted are coding indels found in a human with no major diseases. The coding indels occurr frequently at the N- and C-termini of proteins. These may be functionally neutral because for indels at the end of the protein, most of the protein has already been translated. Indels at the beginning of the protein may be rescued by a downstream translation start site. Hence, using the indel location may help you distinguish between neutral and deleterious indels.
[ Reference]
Transcript Visualization
Placing the cursor over the contents of this column displays a visualization of the transcript in the following format:
<---{}--{}[]--[*.]--[]--[]--[]--[]--[]--[]--[]{}---|
The above example visualization mimics the structure of the transcript containing the indel.
<--- indicates the 3' end
---| indicates the 5' end
{} indicate UTR
[] indicates a coding exon
-- indicats an intron
. indicates the start of insertion or deletion
* indicates the end of deletion
If the 3'end of the transcript appears to the left of the 5' end, as in this case, then the transcript is transcribed from the negative strand.
This transcript has two 3'UTRs, one 5'UTR, nine exons and nine introns.
The indel both starts and ends in the 8th coding exon.
Nucleotide change
The input allele (insertion or deletion) and +/- 5 base pairs are shown. For example,
the user input for insertion variant "10,102760304,102760304,1,GCGGCT" will populate this column with the following information
cggct-GCGGCT-acggc
whereas a user input for deletion variant "12,110521161,110521164,1,/" will populate this column with the following information
TGCTG-ctg-TTGCT
The indel is displayed in red. For insertions, the inserted bases are displayed in uppercase and the flanking bases are displayed in lowercase. For deletions, the deleted bases are displayed in lowercase whereas the flanking bases are displayed in uppercase.
Amino acid change
This column displays the amino acid change caused by the indel. For example
QQTT->QQqTT indicates the addition of amino acid Glutamine ('Q') in the modified protein sequence,
whereas EEeDA->EEDA indicates the deletion of amino acid Glutamic acid, 'E' in the modified protein sequence.
Protein sequence change
This column links to page that contains the original and modified protein sequence files with regions of mismatch (caused due to indel) colored in red. For example, an insertion represented by the user input "1,153108866,153108866,1,CTGCTGCTGCTG" causes an expansion in polyglutamine tract as shown in the following fasta format sequences. The Fasta headers contain the Ensembl transcript ID along with the coordinates of change.
>ENST00000271915; MISMATCH = 80-80
MDTSGHFHDSGVGDLDEDPKCPCPSSGDEQQQQQQQQQQQQPPPPAPPAAPQQPLGPSLQ
PQPPQLQQQQQQQQQQQQQQPPHPLSQLAQLQSQPVHPGLLHSSPTAFRAPPSSNSTAIL
HPSSRQGSQLNLNDHLLGHSPSSTATSGPGGGSRHRQASPLVHRRDSNPFTEIAMSSCKY
SGGVMKPLSRLSASRRNLIEAETEGQPLQLFSPSNPPEIVISSREDNHAHQTLLHHPNAT
HNHQHAGTTASSTTFPKANKRKNQNIGYKLGHRRALFEKRKRLSDYALIFGMFGIVVMVI
ETELSWGLYSKDSMFSLALKCLISLSTIILLGLIIAYHTREVQLFVIDNGADDWRIAMTY
ERILYISLEMLVCAIHPIPGEYKFFWTARLAFSYTPSRAEADVDIILSIPMFLRLYLIAR
VMLLHSKLFTDASSRSIGALNKINFNTRFVMKTLMTICPGTVLLVFSISLWIIAAWTVRV
CERYHDQQDVTSNFLGAMWLISITFLSIGYGDMVPHTYCGKGVCLLTGIMGAGCTALVVA
VVARKLELTKAEKHVHNFMMDTQLTKRIKNAAANVLRETWLIYKHTKLLKKIDHAKVRKH
QRKFLQAIHQLRSVKMEQRKLSDQANTLVDLSKMQNVMYDLITELNDRSEDLEKQIGSLE
SKLEHLTASFNSLPLLIADTLRQQQQQLLSAIIEARGVSVAVGTTHTPISDSPIGVSSTS
FPTPYTSSSSC
>ENST00000271915; MISMATCH = 80-84
MDTSGHFHDSGVGDLDEDPKCPCPSSGDEQQQQQQQQQQQQPPPPAPPAAPQQPLGPSLQ
PQPPQLQQQQQQQQQQQQQQqqqqPPHPLSQLAQLQSQPVHPGLLHSSPTAFRAPPSSNS
TAILHPSSRQGSQLNLNDHLLGHSPSSTATSGPGGGSRHRQASPLVHRRDSNPFTEIAMS
SCKYSGGVMKPLSRLSASRRNLIEAETEGQPLQLFSPSNPPEIVISSREDNHAHQTLLHH
PNATHNHQHAGTTASSTTFPKANKRKNQNIGYKLGHRRALFEKRKRLSDYALIFGMFGIV
VMVIETELSWGLYSKDSMFSLALKCLISLSTIILLGLIIAYHTREVQLFVIDNGADDWRI
AMTYERILYISLEMLVCAIHPIPGEYKFFWTARLAFSYTPSRAEADVDIILSIPMFLRLY
LIARVMLLHSKLFTDASSRSIGALNKINFNTRFVMKTLMTICPGTVLLVFSISLWIIAAW
TVRVCERYHDQQDVTSNFLGAMWLISITFLSIGYGDMVPHTYCGKGVCLLTGIMGAGCTA
LVVAVVARKLELTKAEKHVHNFMMDTQLTKRIKNAAANVLRETWLIYKHTKLLKKIDHAK
VRKHQRKFLQAIHQLRSVKMEQRKLSDQANTLVDLSKMQNVMYDLITELNDRSEDLEKQI
GSLESKLEHLTASFNSLPLLIADTLRQQQQQLLSAIIEARGVSVAVGTTHTPISDSPIGV
SSTSFPTPYTSSSSC
Causes Nonsense Mediated Decay
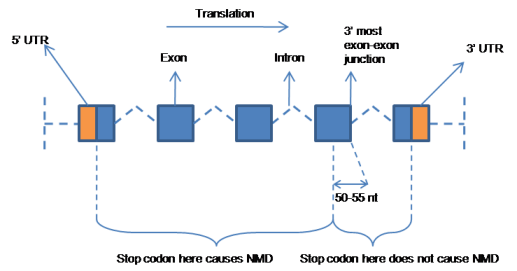
Nonsense mediated decay (NMD) is a cellular mechanism of mRNA surveillance to detect nonsense mutations and prevent the expression of truncated or erroneous proteins [More].
This column indicates whether the input indel is likely to cause NMD. If NMD occurs, then the indel is equivalent to a gene deletion because the mRNA is never translated.
There is no NMD when:
1) the resulting premature termination codon is in the last exon
-or-
2) the resulting premature termintion codon is in the last 50 nucleotides in the second to last exon
[Reference]
Repeat detected
This column gets populated if the input insertion/deletion is found to expand or contract a coding repeat region. For example, an input row '1,153108866,153108866,1,CTGCTGCTGCTG' causes an insertion resulting in the expansion of a poly-glutamine repeat. A poly-glutamine repeat of length 14 that expands to length 18 is illustrated in this column by 'PQL(q)14P-->PQL(q)18P'. The repeat amino acid(s) are shown in parenthesis followed by the repeat number and bounded by flanking amino acids.
Warning: NCBI reference miscall
If you receive a reference miscall warning in the coordinates column (first column) of the output table, this means that your input coordinates overlap or contain a location that is not a true indel, but likely to be an error in NCBI human genome reference sequence.
[Reference]

